We are interested in studying the molecular and cellular mechanisms driving pathogenic processes in the human brain, but also to identify/validate new therapeutic targets or molecules. More specifically, we propose to develop novel approaches to better understand/treat Alzheimer’s disease on the one hand, and brain tumours on the other. To tackle such challenges, we focus on developing advanced in vitro / in cellulo models that are able to recapitulate the disease phenotypes associated to these diseases, and then exploit them to reveal the pathogenic mechanisms at stake. To this end, we use human induced pluripotent stem cells (hiPSCs) and genome editing-based strategies with CRISPR/Cas9 system. Ultimately, we anticipate that our research will lead to the generation of custom-tailored cells and models for treating and understanding brain diseases through manipulation of defined genes and pathways.
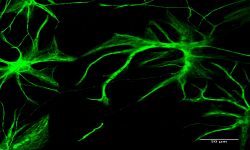
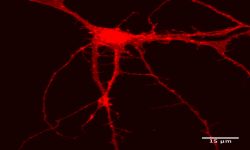
Innovative models to study human diseases are especially crucial for tissues/organs that are largely inaccessible, such as the brain. Due to the overall inaccessibility of human brain cells for laboratory research, brain diseases have largely been studied through the use of simplified cellular models and animal models. Even though large strides have been achieved in our understanding of brain diseases with such models, they also present drawbacks that limit succes when translating promising laboratory findings into the clinic. Our research aims at reducing this gap between laboratory research and translational research by developing human models based on patient-derived iPSCs, which bear the genetic information of the patients and allow differentiating and studying brain cells (e.g., neurons or astrocytes) that will present disease-relevant phenotypes. Of outmost interest, hiPSC-based models are accessible for genetic manipulation and amenable to drug development/screening/testing, which facilitates molecular and cellular studies on disease-affected human cell types and offer a unique opportunity to accelerate investigations for new therapeutic developments.
The "Stem cells, Disease modeling and Neuroregeneration" team is a double edge sword research group that aims at generating pertinent modeling platforms of brain diseases to enhance our fundamental understanding as well as investigate novel therapeutic strategies.
Examining pathogenic mechanisms underlying Alzheimer’s disease
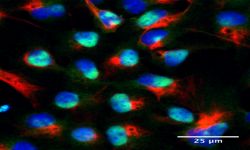
Our goal is to examine the pathogenic mechanisms underlying Alzheimer’s disease (AD) initiation/progression and investigate potential therapeutic avenues by targeting newly identified molecular candidates at play in the disease. To this end, we are using patient-specific hiPSCs presenting mutations (or specific genetic risk factors) in disease-associated genes (e.g., APP, APOE). In addition, we are also generating isogenic sets of hiPSC lines; i.e. either by inserting disease-specific mutations in a control “non-AD” genetic background or by genome editing of disease-associated genetic mutations/risk factors. From those hiPSC lines we are applying differentiation strategies in order to generate different cell types (e.g., neurons and astrocytes) and implement in vitro model systems recapitulating disease-relevant phenotypes found in AD, and then use them to reveal cellular and molecular disease mechanisms. Among the different questions we are investigating with these models, one of our major interests is to study the role of astrocytes in AD pathogenesis as we believe that the crosstalk between neurons and astrocytes is a pivotal determinant in AD through the modulation of factors at the interplay between amyloidosis and neuroinflammation. In addition, and in collaboration with Dr. Santiago Rivera’s team, we use our models to investigate in human settings new molecular targets first identified in mouse models, and evaluate whether they could represent good candidates for the development of therapeutics against AD.
Investigating gliomagenesis with human iPSCs
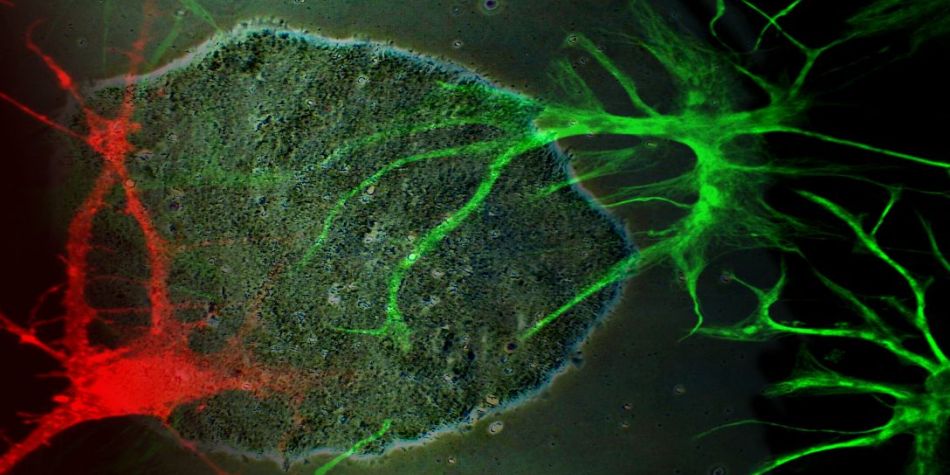
We previously demonstrated that hiPSCs may represent a useful tool for modeling adult gliomagenesis and Glioma tumor initiating cell (GTIC) formation (Sancho-Martinez et al., Nature Communications 2016). The strategy we developed allows for the generation of large collections of patient-specific “induced”-GTICs (iGTICs) bearing different genetic mutations. To follow up on this strategy we are pursuing the development of hiPSC-based models to study gliomagenesis (monolayer-based systems and 3D “brain organoïdes”). Combining our hiPSC-based modeling approaches together with gene-editing technologies, we are examining the role that certain mutations may have on brain cancer predisposition/malignancy and how they synergize during gliomagenesis by controlling tumorigenic mechanisms. This work is, in part, led in collaboration with Pr. Dominique Figarella-Branger’s team. We are also interested in using our in vitro model systems for the screening/testing/validation of novel anti-glioma drug candidates. Along this line, we have an ongoing collaboration with Dr. Michel khrestchatisky’s team and the Vect-Horus company in order to develop a modeling platform mimicking the neuro-glial-vascular unit, with the ultimate goal to provide a useful tool for the screening of compounds and the development of peptide vectors conjugated to newly identified drugs against glioma cells (primarily against GTICs).