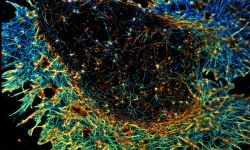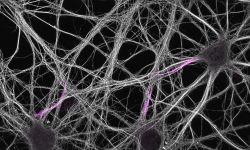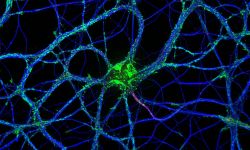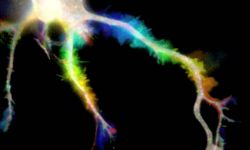

The team started in 2017 as part of the CNRS ATIP program. At NeuroCyto, we are neuronal cell biologists: we want to understand how neurons are organized at the cellular level.
How do neurons differentiate, then build and maintain their complex arborization? How do they establish and conserve their polarity, with the axon and dendrites allowing to send and receive signals? Numerous processes contribute to this organization: elaboration of the cell architecture (thanks to the cytoskeleton), protein transport inside the cell (with diffusion and motor proteins), segregation into distinct compartments (such as axon, synapses, dendritic spines…).
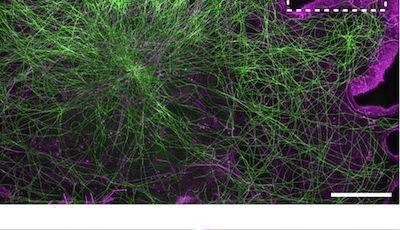
We apply advanced microscopy techniques to directly observe molecular assemblies at the nanoscale in neurons, revealing how they organize the neuron and shape its physiology.
“NeuroCyto: the neuronal cytoskeleton in physiology and disease” is a team that was created in 2017, as part of the Neuropathophysiology Institute (INP, CNRS-Aix Marseille University UMR 7051) in beautiful Marseille, France. The team currently has six members, and we welcome trainees all year long. Motivated students are always welcome to contact us! We aim at building a thriving team to make the best possible science by nurturing openness, exchange, and the excitement of discoveries big and small. At NeuroCyto, we want to understand how neurons are organized at the cellular level. How do they differentiate, then build and maintain their complex arborization? How do they establish and conserve their polarity, with the axon and dendrites allowing to send and receive signals? Numerous processes contribute to this organization: elaboration of the cell architecture (thanks to the cytoskeleton), protein transport inside the cell (with diffusion and motor proteins), segregation into distinct compartments (such as an axon, synapses, dendritic spines…). The NeuroCyto team applies advanced microscopy techniques to directly observe molecular assemblies at the nanoscale level in neurons, revealing how they organize the neuron and shape its physiology.
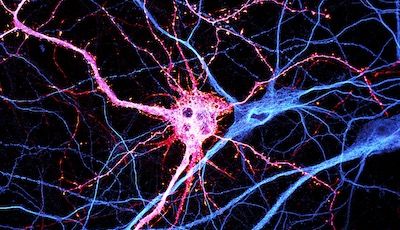
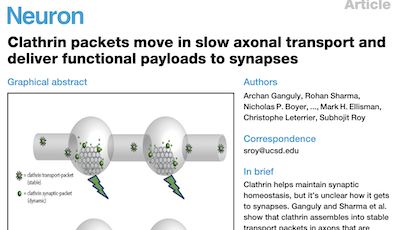
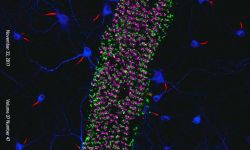
We are currently focusing on actin organization within axons. Several new axonal actin structures have been discovered by us and others, including submembrane actin rings and intra-axonal actin hotspots and trails. We want to understand the functions of these structures and their relevance for physiological processes such as axonal transport and proper functioning of presynaptic boutons. To do so, we take advantage of the beautiful model of hippocampal neurons in low-density culture, so we can visualize the intricate morphology of individual neurons and follow individual axons. We combine live-cell imaging, targeted manipulations and super-resolution microscopy to connect the dynamic behavior of axonal components to the nanoscale organization of the cytoskeleton, and we devote substantial efforts to implement innovative strategies for the labeling, observation and analysis of these processes. We explore the pathophysiological relevance of our finding by studying cellular models of Alzheimer’s disease, as axonal transport and presynaptic function are compromised in the early stages of the disease.
Role of actin-based nanostructures in axons
Following the identification of new axonal actin structures (rings, hotspots and trails), we want to understand their molecular architecture, assembly mechanism and potential interplay. This is done thanks to a combination of live-cell imaging and super-resolution microscopy techniques as well as innovative strategies to selectively perturb actin within axons.
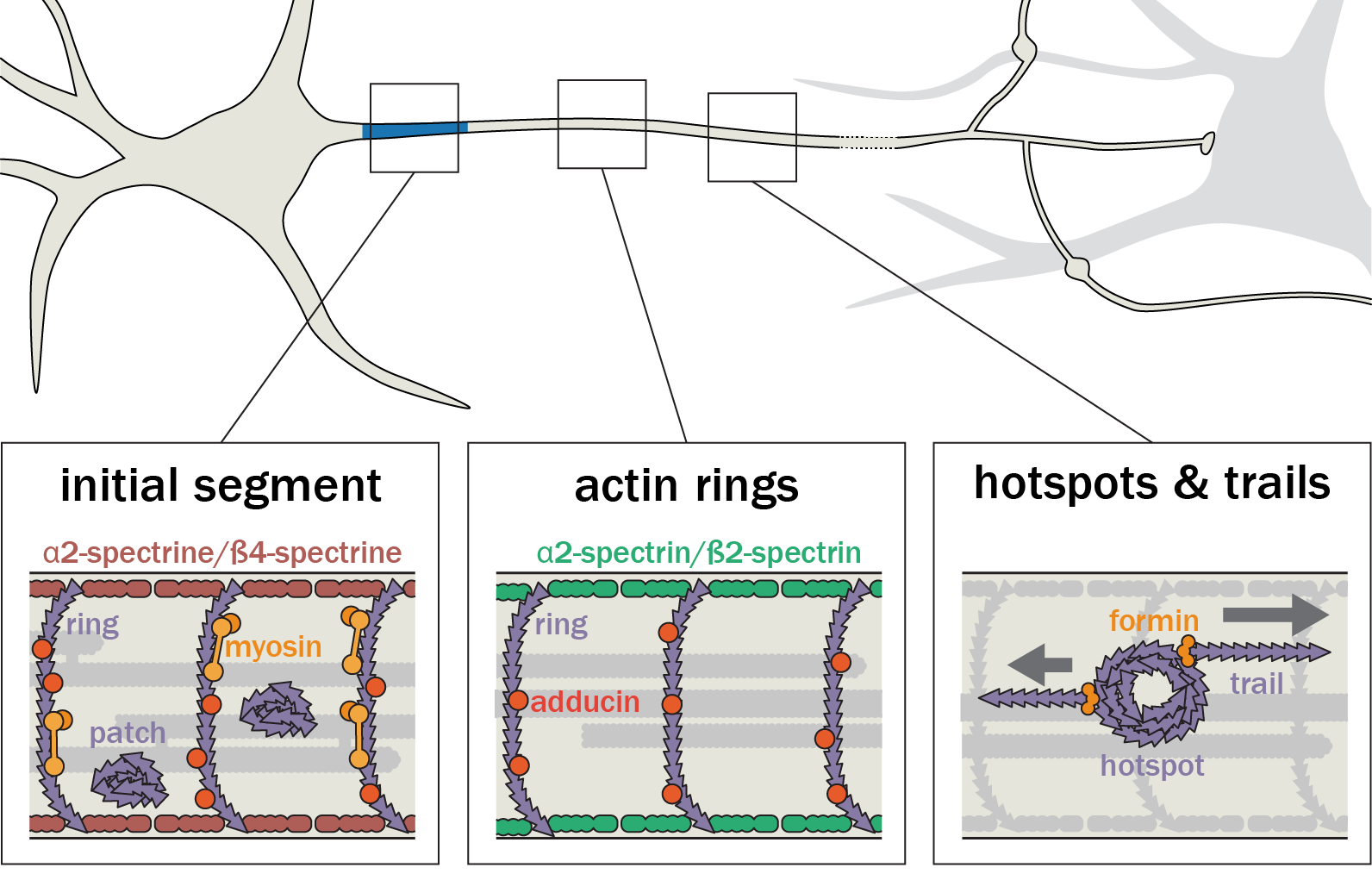
New approaches for labeling, imaging and analysis in super-resolution microscopy
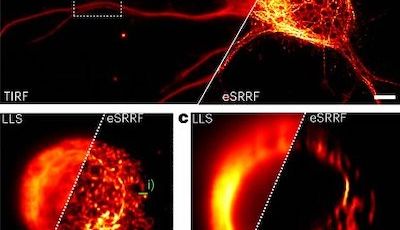
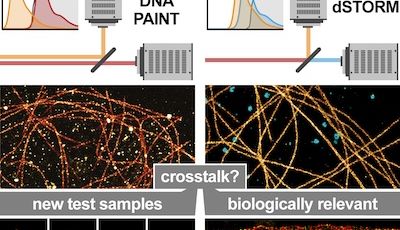
To reveal the organization of the axon, an intricate cellular compartment with extensive length and branching, we use optical super-resolution microscopy. We are specialists of Single Molecule Localization Microscopy (SMLM), and we keep working to push the methods, implementing and testing new techniques as soon as they appear. This includes labeling methods for highly-multiplexed acquisition such as DNA-PAINT, automated acquisition strategies, and new image analysis algorithms.
Cell biology of Alzheimer’s disease in relevant cellular models
Impairment of axonal transport and presynaptic release are early-stage events in Alzheimer’s disease. Using our cell biology background, we want to gain a better understanding of why this happens by studying the nanoscale organization of the axon in Alzheimer’s disease models. Starting with classical rodent models, we want to turn to human models thanks to neurons obtained from induced pluripotent stem cells developed by the Nivet team. These models are still poorly characterized at the subcellular organization level, and it will be interesting to look for specific defects in neurons derived from Alzheimer’s disease patients